Characterization of the epitranscriptomic landscape of HIV-infected cell
In this study, we used a productive HIV infection model, consisting of the CD4+ SupT1 T cell line infected with a VSV-G pseudotyped HIVeGFP-based vector, to explore the transcriptomic and m6A/m5C epitranscriptomic lansdcape upon HIV infection, and compare it to mock-treated cells at 12h, 24h, and 36h post infection.
For more information look at our paper
Experimental design

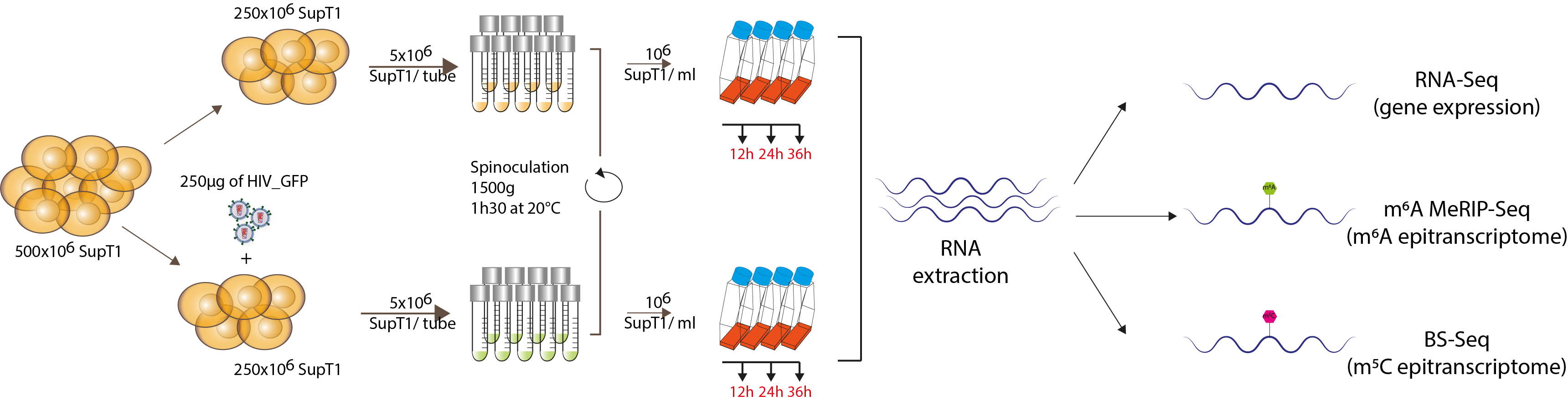
Figure 1. Dynamic analysis of HIV-infected cells. SupT1 cells were either infected with 1 µg/106 cells p24 equivalent of HIV_GFP virus or left uninfected, divided into aliquots of 5*106 cells/ tube and spinoculated for 90 minutes at 1500 g and 20°C to allow nearly universal infection. Cells were then resuspended at a concentration of 106 cells/ml and further incubated. At 12, 24 and 36h post–infection, RNA was extracted from each sample and processed for RNA-Seq to investigate gene expression, MeRIP-Seq and BS-Seq to investigate m6A and m5C epitranscriptomic marks.
MeRIP-Seq: Methylated RNA Immuno-precipitation followed by sequencing; BS-Seq: Bisulfite conversion followed by sequencing. (Detailed protocol in the full text paper)
HI-TEAM
We developed HI-TEAM (HIV-Infected cell Transcriptome and EpitrAnscriptoMe), a user-friendly querying platform based on the iSEE interactive interface [iSEE] to explore all data or any gene of interest.
[iSEE] Rue-Albrecht K, Marini F, Soneson C, Lun ATL (2018). “iSEE: Interactive SummarizedExperiment Explorer.” F1000Research, 7, 741. doi: 10.12688/f1000research.14966.1.
This web resource will allow looking at:
· Genes differentially expressed upon infection
· Genes differentially m6A methylated upon infection
· Genes differentially m5C methylated upon infection